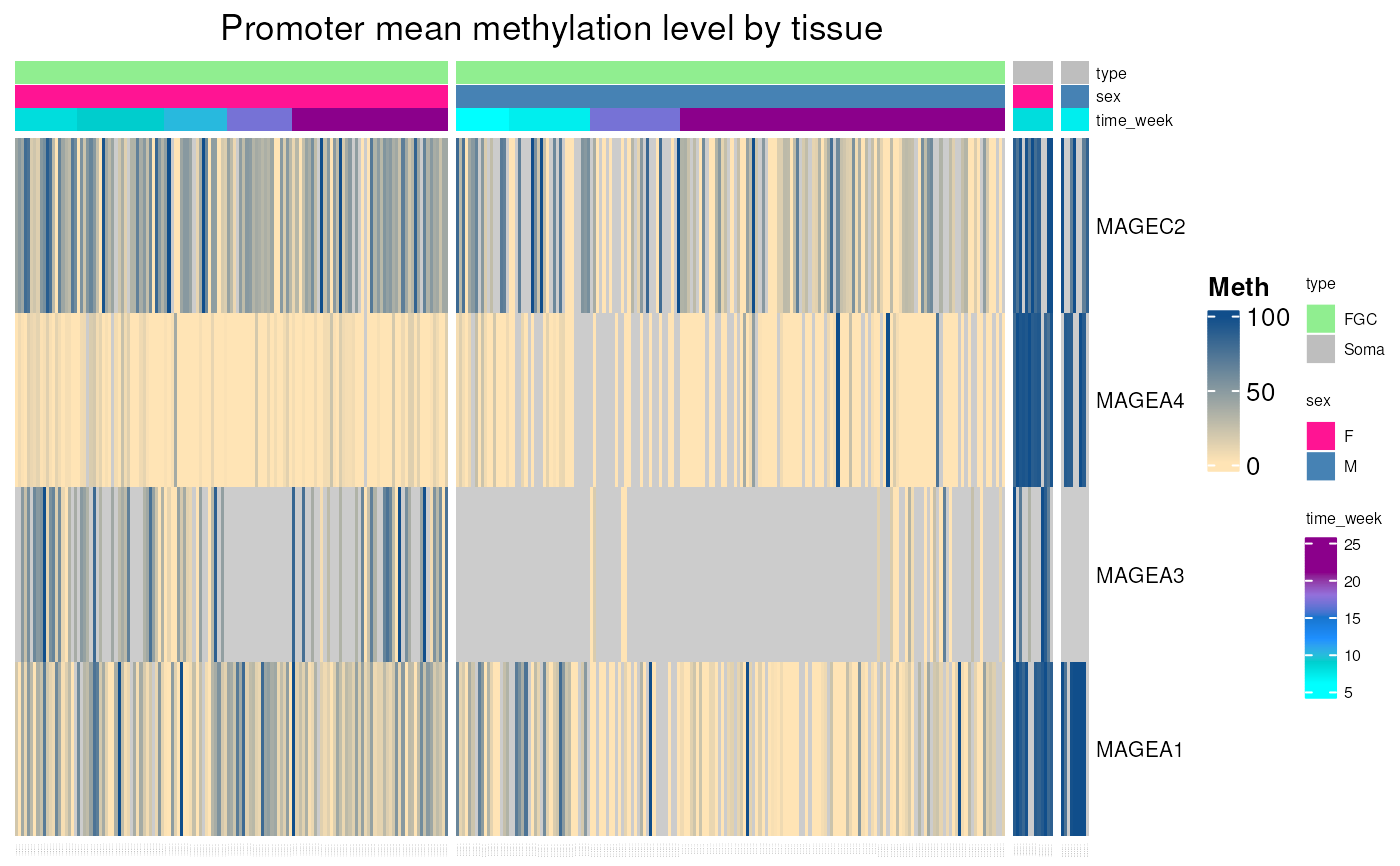
Promoter methylation of any gene in fetal germ cells
Source:R/fetal_germcells_mean_methylation.R
fetal_germcells_mean_methylation.RdPlots a heatmap of mean promoter methylation levels of any genes in fetal germ cells, using WGSB data from "Dissecting the epigenomic dynamics of human fetal germ cell development at single-cell resolution" (Li et al. 2021). Methylation levels in tissues correspond to the mean methylation of CpGs located in range of 1000 pb upstream and 500 pb downstream from gene TSS.
Arguments
- genes
characternaming the selected genes. The default value,NULL, takes all CT (specific) genes.- include_CTP
logical(1)IfTRUE, CTP genes are included. (FALSEby default).- values_only
logical(1),FALSEby default. IfTRUE, the function will return the methylation values in all samples instead of the heatmap.
Value
Heatmap of mean promoter methylation of any gene in normal tissues.
If values_only = TRUE, a SummarizeExperiment with methylation values is
returned instead.
Examples
fetal_germcells_mean_methylation()
#> see ?CTdata and browseVignettes('CTdata') for documentation
#> loading from cache
#> Warning: 2 out of 146 names invalid: FAM230C, FLJ36000.
#> See the manual page for valid types.
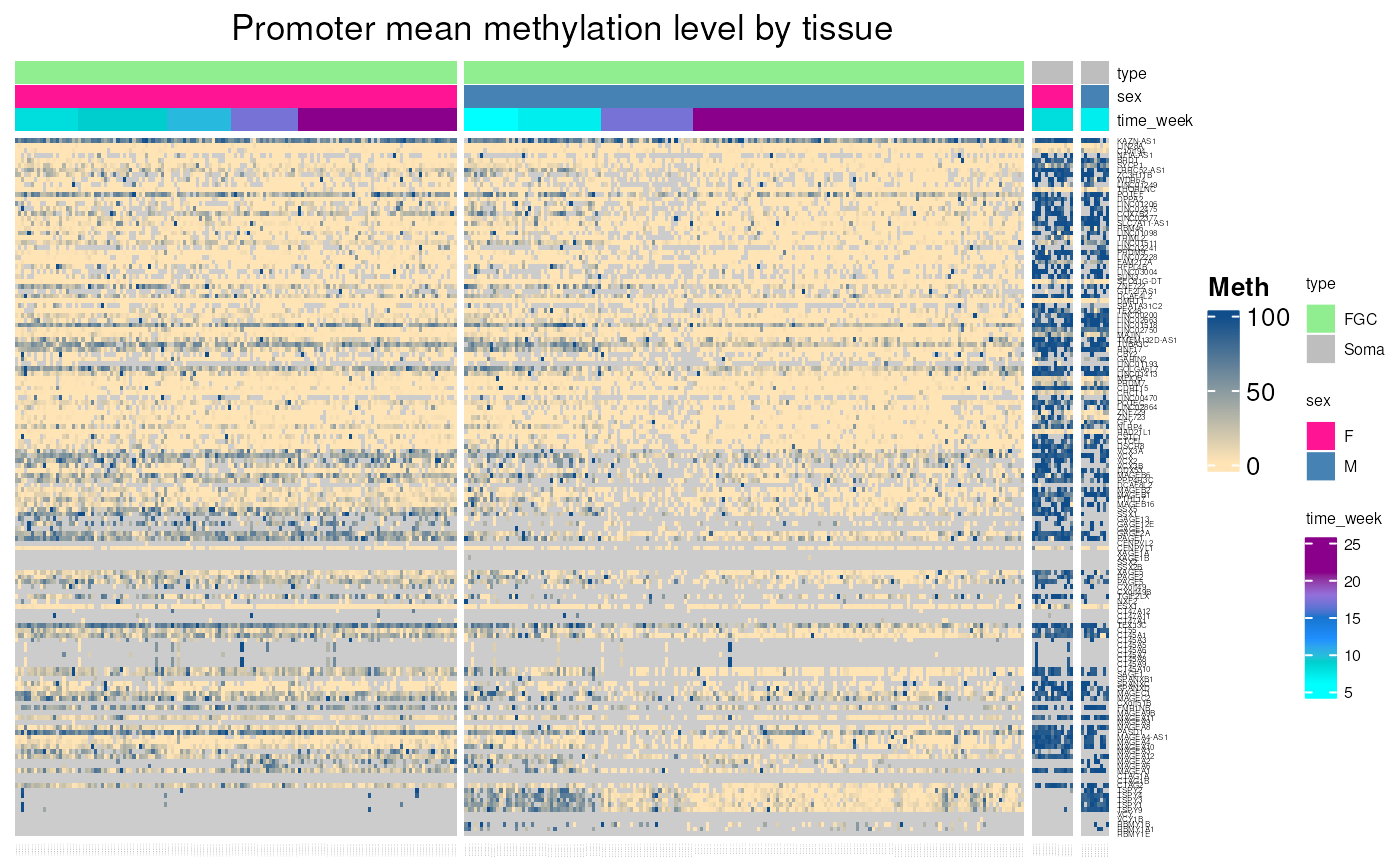 fetal_germcells_mean_methylation(c("MAGEA1", "MAGEA3", "MAGEA4", "MAGEC2"))
#> see ?CTdata and browseVignettes('CTdata') for documentation
#> loading from cache
fetal_germcells_mean_methylation(c("MAGEA1", "MAGEA3", "MAGEA4", "MAGEC2"))
#> see ?CTdata and browseVignettes('CTdata') for documentation
#> loading from cache