Chapter 7 Joining tables
Learning Objectives:
Understand the need and concept of table joins
Understand the different types of joins
Understand the importance of keys in joins and the implications of using non-unique keys
In many real life situations, data are spread across multiple tables. Usually this occurs because different types of information about a subject, e.g. a patient, are collected from different sources.
It may be desirable for some analyses to combine data from two or more tables into a single data frame based on a column that would be common to all the tables, for example, an attribute that uniquely identifies the subjects.
The dplyr package provides a set of join functions for combining
two data frames based on matches within specified columns.
For further reading, please refer to the chapter about table joins in Grolemund and Wickham (2017)Grolemund, Garrett, and Hadley Wickham. 2017. R for Data Science. O’Reilly Media. https://r4ds.had.co.nz/..
The Data Transformation Cheat Sheet also provides a short overview on table joins.
7.1 Combining tables
We are going to illustrate join using a common example from the bioinformatics world, where annotations about genes are scattered in different tables that have one or more shared columns.
The data we are going to use are available in the following package.
# install.packages(c("BiocManager", "remotes"))
# BiocManager::install("UCLouvain-CBIO/rWSBIM1207")
library("rWSBIM1207")##
## This is 'rWSBIM1207' version 0.1.14data(jdf)The data is composed of several tables.
The first table, jdf1, contains protein UniProt9 UniProt is the protein information database. Its mission is to provide the scientific community with a comprehensive, high-quality and freely accessible resource of protein sequence and functional information. unique
accession number (uniprot variable), the most likely sub-cellular
localisation of these respective proteins (organelle variable) as
well as the proteins identifier (entry).
jdf1## # A tibble: 25 × 3
## uniprot organelle entry
## <chr> <chr> <chr>
## 1 P26039 Actin cytoskeleton TLN1_MOUSE
## 2 Q99PL5 Endoplasmic reticulum/Golgi apparatus RRBP1_MOUSE
## 3 Q6PB66 Mitochondrion LPPRC_MOUSE
## 4 P11276 Extracellular matrix FINC_MOUSE
## 5 Q6PR54 Nucleus - Chromatin RIF1_MOUSE
## 6 Q05793 Extracellular matrix PGBM_MOUSE
## 7 P19096 Cytosol FAS_MOUSE
## 8 Q9JKF1 Plasma membrane IQGA1_MOUSE
## 9 Q9QZQ1-2 Plasma membrane AFAD_MOUSE
## 10 Q6NS46 Nucleus - Non-chromatin RRP5_MOUSE
## # … with 15 more rowsThe second table, jdf2, contains the name of the gene that codes for the
protein (gene_name variable), a description of the gene
(description variable), the uniprot accession number (this is the
common variable that can be used to join tables) and the species the
protein information comes from (organism variable).
jdf2## # A tibble: 25 × 4
## gene_name description uniprot organism
## <chr> <chr> <chr> <chr>
## 1 Iqgap1 Ras GTPase-activating-like protein IQGAP1 Q9JKF1 Mmus
## 2 Hspa5 78 kDa glucose-regulated protein P20029 Mmus
## 3 Pdcd11 Protein RRP5 homolog Q6NS46 Mmus
## 4 Tfrc Transferrin receptor protein 1 Q62351 Mmus
## 5 Hspd1 60 kDa heat shock protein, mitochondrial P63038 Mmus
## 6 Tln1 Talin-1 P26039 Mmus
## 7 Smc1a Structural maintenance of chromosomes protein 1A Q9CU62 Mmus
## 8 Lamc1 Laminin subunit gamma-1 P02468 Mmus
## 9 Hsp90b1 Endoplasmin P08113 Mmus
## 10 Mia3 Melanoma inhibitory activity protein 3 Q8BI84 Mmus
## # … with 15 more rowsWe now want to join these two tables into a single one containing all variables.
We are going to use the full_join function of dplyr to do
so,
Th function will automatically find the common variable (in this case uniprot)
to match observations from the first and second table.
library("dplyr")
full_join(jdf1, jdf2)## Joining, by = "uniprot"## # A tibble: 25 × 6
## uniprot organelle entry gene_name description organism
## <chr> <chr> <chr> <chr> <chr> <chr>
## 1 P26039 Actin cytoskele… TLN1_M… Tln1 Talin-1 Mmus
## 2 Q99PL5 Endoplasmic ret… RRBP1_… Rrbp1 Ribosome-binding protei… Mmus
## 3 Q6PB66 Mitochondrion LPPRC_… Lrpprc Leucine-rich PPR motif-… Mmus
## 4 P11276 Extracellular m… FINC_M… Fn1 Fibronectin Mmus
## 5 Q6PR54 Nucleus - Chrom… RIF1_M… Rif1 Telomere-associated pro… Mmus
## 6 Q05793 Extracellular m… PGBM_M… Hspg2 Basement membrane-speci… Mmus
## 7 P19096 Cytosol FAS_MO… Fasn Fatty acid synthase Mmus
## 8 Q9JKF1 Plasma membrane IQGA1_… Iqgap1 Ras GTPase-activating-l… Mmus
## 9 Q9QZQ1-2 Plasma membrane AFAD_M… Mllt4 Isoform 1 of Afadin Mmus
## 10 Q6NS46 Nucleus - Non-c… RRP5_M… Pdcd11 Protein RRP5 homolog Mmus
## # … with 15 more rowsIn these examples, each observation of the jdf1 and jdf2
tables are uniquely identified by their UniProt accession number. Such
variables are called keys. Keys are used to match observations
across different tables.
Now let’s look at a third table, jdf3. It also contains the column uniProt,
but it is written diffently!
jdf3## # A tibble: 25 × 4
## gene_name description UniProt organism
## <chr> <chr> <chr> <chr>
## 1 Iqgap1 Ras GTPase-activating-like protein IQGAP1 Q9JKF1 Mmus
## 2 Hspa5 78 kDa glucose-regulated protein P20029 Mmus
## 3 Pdcd11 Protein RRP5 homolog Q6NS46 Mmus
## 4 Tfrc Transferrin receptor protein 1 Q62351 Mmus
## 5 Hspd1 60 kDa heat shock protein, mitochondrial P63038 Mmus
## 6 Tln1 Talin-1 P26039 Mmus
## 7 Smc1a Structural maintenance of chromosomes protein 1A Q9CU62 Mmus
## 8 Lamc1 Laminin subunit gamma-1 P02468 Mmus
## 9 Hsp90b1 Endoplasmin P08113 Mmus
## 10 Mia3 Melanoma inhibitory activity protein 3 Q8BI84 Mmus
## # … with 15 more rowsIn case none of the variable names match, we can set manually the variables to
use for the matching. These variables can be set using the by argument,
as shown below with the jdf1 (as
above) and jdf3 tables, where the UniProt accession number is
encoded using a different capitalisation.
names(jdf3)## [1] "gene_name" "description" "UniProt" "organism"full_join(jdf1, jdf3, by = c("uniprot" = "UniProt"))## # A tibble: 25 × 6
## uniprot organelle entry gene_name description organism
## <chr> <chr> <chr> <chr> <chr> <chr>
## 1 P26039 Actin cytoskele… TLN1_M… Tln1 Talin-1 Mmus
## 2 Q99PL5 Endoplasmic ret… RRBP1_… Rrbp1 Ribosome-binding protei… Mmus
## 3 Q6PB66 Mitochondrion LPPRC_… Lrpprc Leucine-rich PPR motif-… Mmus
## 4 P11276 Extracellular m… FINC_M… Fn1 Fibronectin Mmus
## 5 Q6PR54 Nucleus - Chrom… RIF1_M… Rif1 Telomere-associated pro… Mmus
## 6 Q05793 Extracellular m… PGBM_M… Hspg2 Basement membrane-speci… Mmus
## 7 P19096 Cytosol FAS_MO… Fasn Fatty acid synthase Mmus
## 8 Q9JKF1 Plasma membrane IQGA1_… Iqgap1 Ras GTPase-activating-l… Mmus
## 9 Q9QZQ1-2 Plasma membrane AFAD_M… Mllt4 Isoform 1 of Afadin Mmus
## 10 Q6NS46 Nucleus - Non-c… RRP5_M… Pdcd11 Protein RRP5 homolog Mmus
## # … with 15 more rowsAs can be seen above, the variable name of the first table is retained in the joined one.
► Question
Using the full_join function, join tables jdf4
and jdf5. What has happened for observations P26039 and P02468?
► Solution
7.2 Different types of joins
Above, we have used the full_join function, that fully joins two
tables and keeps all observations, adding missing values if
necessary. Sometimes, we want to be selective, and keep observations
that are present in only one or both tables.
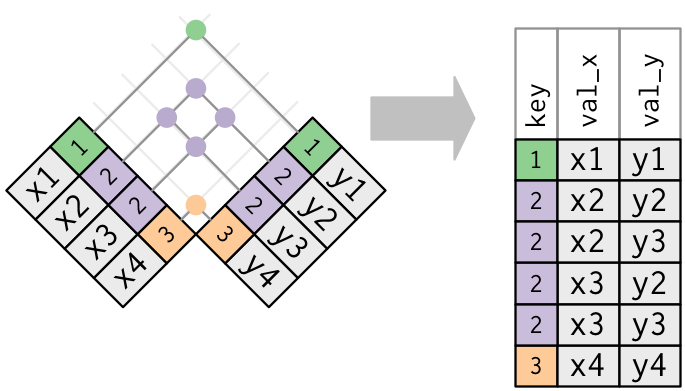
- An inner join keeps observations that are present in both tables.
Figure 7.1: An inner join matches pairs of observation matching in both tables, this dropping those that are unique to one table. Figure taken from R for Data Science.
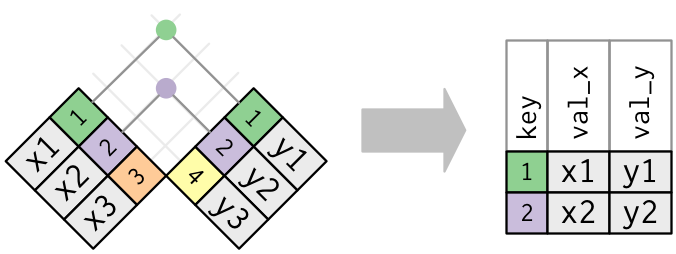
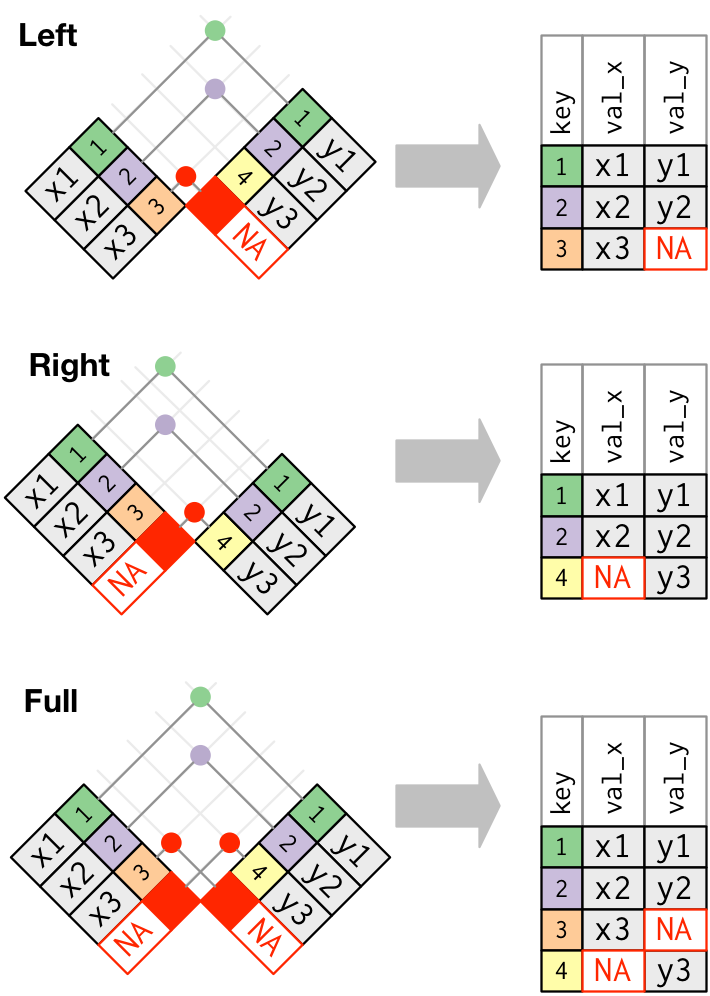
- A left join keeps observations that are present in the left (first) table, dropping those that are only present in the other.
- A right join keeps observations that are present in the right (second) table, dropping those that are only present in the other.
- A full join keeps all observations.
Figure 7.2: Outer joins match observations that appear in at least on table, filling up missing values with NA values. Figure taken from R for Data Science.
► Question
Join tables jdf4 and jdf5, keeping only observations in jdf4.
► Solution
► Question
Join tables jdf4 and jdf5, keeping only observations in jdf5.
► Solution
► Question
Join tables jdf4 and jdf5, keeping observations observed in both tables.
► Solution
7.3 Multiple matches
Sometimes, keys aren’t unique.
In the jdf6 table below, we see that the accession number Q99PL5 is repeated
twice. According to this table, the ribosomial protein binding protein 1 localises in the
endoplasmic reticulum and in the Golgi apparatus.
jdf6## # A tibble: 5 × 4
## uniprot organelle entry isoform
## <chr> <chr> <chr> <dbl>
## 1 P26039 Actin cytoskeleton TLN1_MOUSE 1
## 2 Q99PL5 Endoplasmic reticulum RRBP1_MOUSE 1
## 3 Q99PL5 Golgi apparatus RRBP1_MOUSE 2
## 4 Q6PB66 Mitochondrion LPPRC_MOUSE 1
## 5 P11276 Extracellular matrix FINC_MOUSE 1If we now want to join jdf6 and jdf2, the variables of the latter
will be duplicated.
inner_join(jdf6, jdf2)## Joining, by = "uniprot"## # A tibble: 5 × 7
## uniprot organelle entry isoform gene_name description organism
## <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
## 1 P26039 Actin cytoskeleton TLN1_MOUSE 1 Tln1 Talin-1 Mmus
## 2 Q99PL5 Endoplasmic reticulum RRBP1_MOUSE 1 Rrbp1 Ribosome-b… Mmus
## 3 Q99PL5 Golgi apparatus RRBP1_MOUSE 2 Rrbp1 Ribosome-b… Mmus
## 4 Q6PB66 Mitochondrion LPPRC_MOUSE 1 Lrpprc Leucine-ri… Mmus
## 5 P11276 Extracellular matrix FINC_MOUSE 1 Fn1 Fibronectin MmusIn the case above, repeating is useful, as it completes jdf6 with
correct information from jdf2.
But one needs however to be careful when duplicated keys exist in both tables.
Let’s now use jdf7 for the join. It also has 2 entries for the uniprot Q99PL5
jdf7## # A tibble: 5 × 6
## gene_name description uniprot organism isoform_num measure
## <chr> <chr> <chr> <chr> <dbl> <dbl>
## 1 Rrbp1 Ribosome-binding protein 1 Q99PL5 Mmus 1 102
## 2 Rrbp1 Ribosome-binding protein 1 Q99PL5 Mmus 2 3
## 3 Iqgap1 Ras GTPase-activating-like pro… Q9JKF1 Mmus 1 13
## 4 Hspa5 78 kDa glucose-regulated prote… P20029 Mmus 1 54
## 5 Pdcd11 Protein RRP5 homolog Q6NS46 Mmus 1 28Let’s we create an inner join between jdf6 and jdf7 (both having duplicated Q99PL5 entries)
inner_join(jdf6, jdf7)## Joining, by = "uniprot"## # A tibble: 4 × 9
## uniprot organelle entry isoform gene_name description organism isoform_num
## <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <dbl>
## 1 Q99PL5 Endoplasmi… RRBP1… 1 Rrbp1 Ribosome-bi… Mmus 1
## 2 Q99PL5 Endoplasmi… RRBP1… 1 Rrbp1 Ribosome-bi… Mmus 2
## 3 Q99PL5 Golgi appa… RRBP1… 2 Rrbp1 Ribosome-bi… Mmus 1
## 4 Q99PL5 Golgi appa… RRBP1… 2 Rrbp1 Ribosome-bi… Mmus 2
## # … with 1 more variable: measure <dbl>► Question
Interpret the result of the inner join above, where both tables have duplicated keys.
► Solution
7.4 Matching across multiple keys
So far, we have matched tables using a single key (possibly with different names in the two tables). Sometimes, it is necessary to match tables using multiple keys. A typical example is when multiple variables are needed to discriminate different rows in a tables.
Following up from the last example, we see that the duplicated UniProt
accession numbers in the jdf6 and jdf7 tables refer to different
isoforms of the same RRBP1 gene.
jdf6## # A tibble: 5 × 4
## uniprot organelle entry isoform
## <chr> <chr> <chr> <dbl>
## 1 P26039 Actin cytoskeleton TLN1_MOUSE 1
## 2 Q99PL5 Endoplasmic reticulum RRBP1_MOUSE 1
## 3 Q99PL5 Golgi apparatus RRBP1_MOUSE 2
## 4 Q6PB66 Mitochondrion LPPRC_MOUSE 1
## 5 P11276 Extracellular matrix FINC_MOUSE 1jdf7## # A tibble: 5 × 6
## gene_name description uniprot organism isoform_num measure
## <chr> <chr> <chr> <chr> <dbl> <dbl>
## 1 Rrbp1 Ribosome-binding protein 1 Q99PL5 Mmus 1 102
## 2 Rrbp1 Ribosome-binding protein 1 Q99PL5 Mmus 2 3
## 3 Iqgap1 Ras GTPase-activating-like pro… Q9JKF1 Mmus 1 13
## 4 Hspa5 78 kDa glucose-regulated prote… P20029 Mmus 1 54
## 5 Pdcd11 Protein RRP5 homolog Q6NS46 Mmus 1 28To uniquely identify isoforms, we should consider two keys:
the UniProt accession number (named
uniprotin both tables)and the isoform number (called
isoformandisoform_numrespectively)
Because the isoform status was encoded using different names
(which is, of course a source of confusion), jdf6 and jdf7 are
only automatically joined based on the shared uniprot key.
If the isoform status was encoded the same way in both tables, the join would have been automatically done on both keys!
Here, we need to join using both keys and need to explicitly name the variables used for the join.
inner_join(jdf6, jdf7, by = c("uniprot" = "uniprot", "isoform" = "isoform_num"))## # A tibble: 2 × 8
## uniprot organelle entry isoform gene_name description organism measure
## <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <dbl>
## 1 Q99PL5 Endoplasmic… RRBP1_… 1 Rrbp1 Ribosome-bind… Mmus 102
## 2 Q99PL5 Golgi appar… RRBP1_… 2 Rrbp1 Ribosome-bind… Mmus 3We now see that isoform 1 localised to the ER and has a measured value of 102, while isoform 2, that localised to the GA, has a measured value of 3.
Ideally, the isoform variables should be named identically in the two tables to enable an automatic join with the two keys.
An alternative could be to rename the isoform_num from jdf7 in order to
have the both keys names present in both tables, enabling an automatic join.
This can be done easily using the rename function from dplyr package.
jdf7 %>% rename(isoform = isoform_num)## # A tibble: 5 × 6
## gene_name description uniprot organism isoform measure
## <chr> <chr> <chr> <chr> <dbl> <dbl>
## 1 Rrbp1 Ribosome-binding protein 1 Q99PL5 Mmus 1 102
## 2 Rrbp1 Ribosome-binding protein 1 Q99PL5 Mmus 2 3
## 3 Iqgap1 Ras GTPase-activating-like protein… Q9JKF1 Mmus 1 13
## 4 Hspa5 78 kDa glucose-regulated protein P20029 Mmus 1 54
## 5 Pdcd11 Protein RRP5 homolog Q6NS46 Mmus 1 28inner_join(jdf6,
jdf7 %>%
rename(isoform = isoform_num)) ## Joining, by = c("uniprot", "isoform")## # A tibble: 2 × 8
## uniprot organelle entry isoform gene_name description organism measure
## <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <dbl>
## 1 Q99PL5 Endoplasmic… RRBP1_… 1 Rrbp1 Ribosome-bind… Mmus 102
## 2 Q99PL5 Golgi appar… RRBP1_… 2 Rrbp1 Ribosome-bind… Mmus 37.5 Row and column binding
There are two other important functions in R, rbind and cbind,
that can be used to combine two dataframes.
-
cbindcan be used to bind two data frames by columns, but both must have the same number of rows.
d2## a b
## 1 4 4
## 2 5 5d3## v1 v2 v3
## 1 1 3 5
## 2 2 4 6cbind(d2, d3)## a b v1 v2 v3
## 1 4 4 1 3 5
## 2 5 5 2 4 6-
rbind, can be used to bind two data frames by rows, but both must have the same number of columns, and the same column names!
d1## x y
## 1 1 1
## 2 2 2
## 3 3 3d2## a b
## 1 4 4
## 2 5 5using rbind(d1, d2) would produce an error because both data frames do not have the
same column names (even if they have the same number of columns)
If we change the names of d2, it works!
names(d2) <- names(d1)
d1## x y
## 1 1 1
## 2 2 2
## 3 3 3d2## x y
## 1 4 4
## 2 5 5rbind(d1, d2)## x y
## 1 1 1
## 2 2 2
## 3 3 3
## 4 4 4
## 5 5 5Note that beyond the dimensions and column names that are required to
match, the real meaning of rbind is to bind data frames that contain
observations for the same set of variables - there is more than only
the column names!
Note: rbind and cbind are base R functions. The tidyverse
alternatives from the dplyr package are bind_rows and bind_cols
and work similarly.
Page built: 2021-10-01 using R version 4.1.1 (2021-08-10)